import numpy import pandas from sklearn.feature_selection import SelectKBest from sklearn.feature_selection import chi2 from sklearn.feature_selection import RFE from sklearn.linear_model import LogisticRegression from sklearn.decomposition import PCA from sklearn.ensemble import ExtraTreesClassifier
import matplotlib.pyplot as plt from pandas.plotting import scatter_matrix
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFoldfrom sklearn.tree import DecisionTreeClassifier from sklearn.neighbors import KNeighborsClassifier from sklearn.discriminant_analysis import LinearDiscriminantAnalysis from sklearn.naive_bayes import GaussianNB from sklearn.svm import SVC from sklearn.svm import LinearSVC from sklearn.linear_model import SGDClassifier from sklearn.ensemble import RandomForestClassifier from sklearn.metrics import accuracy_score from sklearn.metrics import mean_squared_error
from keras.models import Sequential from keras.layers import Dense from keras.layers import Dropout from keras.utils import np_utils #from sklearn import preprocessing from sklearn.model_selection import StratifiedKFold from keras.constraints import maxnorm # fix random seed for reproducibility seed = 7 numpy.random.seed(seed)
# load dataset dataframe = pandas.read_csv("./input/ecoli.csv", delim_whitespace=True) # Assign names to Columns dataframe.columns = ['seq_name', 'mcg', 'gvh', 'lip', 'chg', 'aac', 'alm1', 'alm2', 'site'] dataframe = dataframe.drop('seq_name', axis=1) # Encode Data dataframe.site.replace(('cp', 'im', 'pp', 'imU', 'om', 'omL', 'imL', 'imS'),(1,2,3,4,5,6,7,8), inplace=True)
print("Head:", dataframe.head())
print("Statistical Description:", dataframe.describe())
print("Shape:", dataframe.shape) print("Data Types:", dataframe.dtypes)
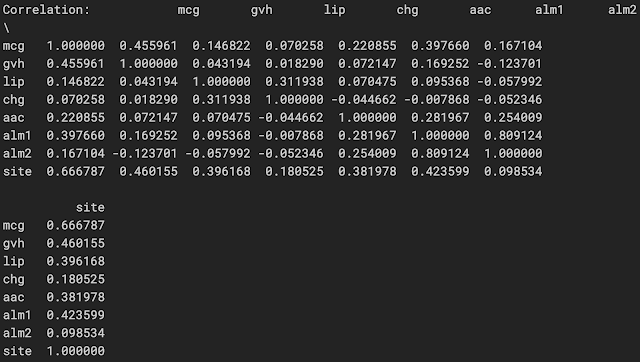
print("Correlation:", dataframe.corr(method='pearson'))
'mcg'(McGeoch的信號序列識別方法)與'site'(蛋白定位位點)具有最高的相關性(這是正相關),其次是'gvh'(von Heijne的信號序列識別方法),它也是 正相關,'alm2'(從序列中排除推定的可切割信號區域後ALOM程序的分數)具有最小的相關性
dataset = dataframe.values X = dataset[:,0:7] Y = dataset[:,7]
print(X.shape)
print(type(X))
print(type(X[0]))
print(type(X[0][0]))
(335, 7)
<class 'numpy.ndarray'>
<class 'numpy.ndarray'>
<class 'numpy.float64'>
#Feature Selection model = LogisticRegression(multi_class='auto', solver='lbfgs') rfe = RFE(model, 3) fit = rfe.fit(X, Y) print("Number of Features: ", fit.n_features_) print("Selected Features: ", fit.support_) print("Feature Ranking: ", fit.ranking_)
'mcg','gvh'和'alm1'(ALOM膜跨越區域預測程序的分數)是使用遞歸特徵消除(Recursive Feature Elimination)來預測“ Income”的前3個選定特徵/特徵組合,第1和2個通常是兩個屬性 與“site”類的最高相關性
plt.hist((dataframe.site))
大多數數據集的樣本都在'cp'(細胞質),'im'(沒有信號序列的內膜)和'pp'(perisplasm)輸出類別的順序內
dataframe.plot(kind='density', subplots=True, layout=(3,4), sharex=False, sharey=False)
除了'mcg'和'aac'之外,大多數屬性都有正偏差(positive skews)
dataframe.plot(kind='box', subplots=True, layout=(3,4), sharex=False, sharey=False)
fig = plt.figure() ax = fig.add_subplot(111) cax = ax.matshow(dataframe.corr(), vmin=-1, vmax=1) fig.colorbar(cax) ticks = numpy.arange(0,7,1) ax.set_xticks(ticks) ax.set_yticks(ticks) ax.set_xticklabels(dataframe.columns) ax.set_yticklabels(dataframe.columns)
'mcg'具有最高的正相關性
num_instances = len(X) models = [] models.append(('LR', LogisticRegression(multi_class='auto', solver='lbfgs'))) models.append(('LDA', LinearDiscriminantAnalysis())) models.append(('KNN', KNeighborsClassifier())) models.append(('CART', DecisionTreeClassifier())) models.append(('NB', GaussianNB())) models.append(('SVM', SVC(gamma="auto"))) models.append(('L_SVM', LinearSVC())) models.append(('ETC', ExtraTreesClassifier(n_estimators=100))) models.append(('RFC', RandomForestClassifier(n_estimators=100))) # Evaluations results = [] names = [] for name, model in models: # Fit the model model.fit(X, Y) predictions = model.predict(X) # Evaluate the model kfold = cross_validation.KFold(n=num_instances, n_folds=10, random_state=seed) cv_results = cross_validation.cross_val_score(model, X, Y, cv=kfold, scoring='accuracy') results.append(cv_results) names.append(name) msg = "%s: %f (%f)" % (name, cv_results.mean(), cv_results.std()) print(msg)
#boxplot algorithm Comparison fig = plt.figure() fig.suptitle('Algorithm Comparison') ax = fig.add_subplot(111) plt.boxplot(results) ax.set_xticklabels(names) plt.show()
'Naive Bayes'和'Linear Discriminant Analysis'是此數據集的最佳估算器/模型,可以進一步探索它們並調整其超參數
# Define 10-fold Cross Valdation Test Harness kfold = StratifiedKFold(n_splits=10, shuffle=True, random_state=seed) cvscores = [] for train, test in kfold.split(X, Y): # create model model = Sequential() model.add(Dense(20, input_dim=7, kernel_initializer='uniform', activation='relu')) model.add(Dropout(0.2)) model.add(Dense(10, kernel_initializer='uniform', activation='relu', kernel_constraint=maxnorm(3))) model.add(Dropout(0.2)) model.add(Dense(5, kernel_initializer='uniform', activation='relu')) model.add(Dense(1, kernel_initializer='uniform', activation='relu')) # Compile model model.compile(loss='mean_squared_error', optimizer='adam', metrics=['accuracy']) # Fit the model model.fit(X[train], Y[train], epochs=200, batch_size=10, verbose=0) # Evaluate the model scores = model.evaluate(X[test], Y[test], verbose=0) print("%s: %.2f%%" % (model.metrics_names[1], scores[1]*100)) cvscores.append(scores[1] * 100) print("%.2f%% (+/- %.2f%%)" % (numpy.mean(cvscores), numpy.std(cvscores)))
參考
https://www.kaggle.com/elikplim/predicting-the-localization-sites-of-proteins















沒有留言:
張貼留言